drLumi: An open-source package to manage data, calibrate, and conduct quality control of multiplex bead-based immunoassays data analysis
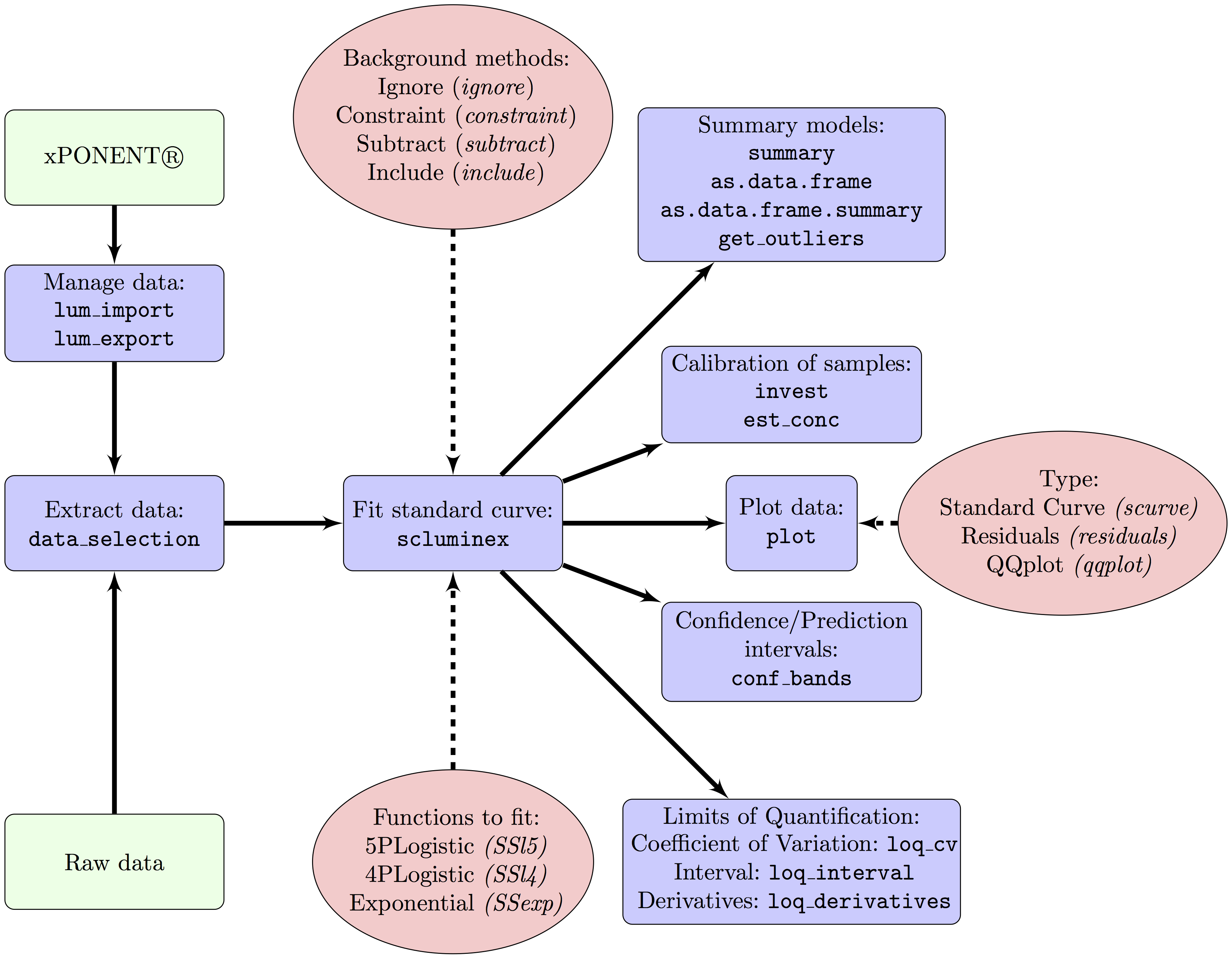
Multiplex bead-based immunoassays are used to measure concentrations of several analytes simultaneously. These assays include control standard curves (SC) to reduce between-plate variability and normalize quantitation of analytes of biological samples. Suboptimal calibration might result in large random error and decreased number of samples with analyte concentrations within the limits of quantification. Suboptimal calibration may be a consequence of poor fitness of the functions used for the SC, the treatment of the background noise and the method used to estimate the limits of quantification. Currently assessment of fitness of curves is largely dependent on operator and that may add additional error. Moreover, there is no software to automate data managing and quality control. In this article we present a R package, drLumi, with functions for managing data, calibrating assays and performing quality control. To optimize the assay the package implements: i) three dose-response functions, ii) four approaches for treating background noise and iii) three methods for estimating limits of quantifications. Other implemented functions are focused on the quality control of the fitted standard curve: detection of outliers, estimation of the confidence or prediction interval, and estimation of summary statistics. With demonstration purpose, we apply the software to 30 cytokines, chemokines and growth factors measured in a multiplex bead-based immunoassay in a study aiming to measure correlates of risk or protection from malaria of the RTS,S malaria vaccine nested in the Phase 3 randomized controlled trial of this vaccine.