EEGNET: An Open Source Tool for Analyzing and Visualizing M/EEG Connectome
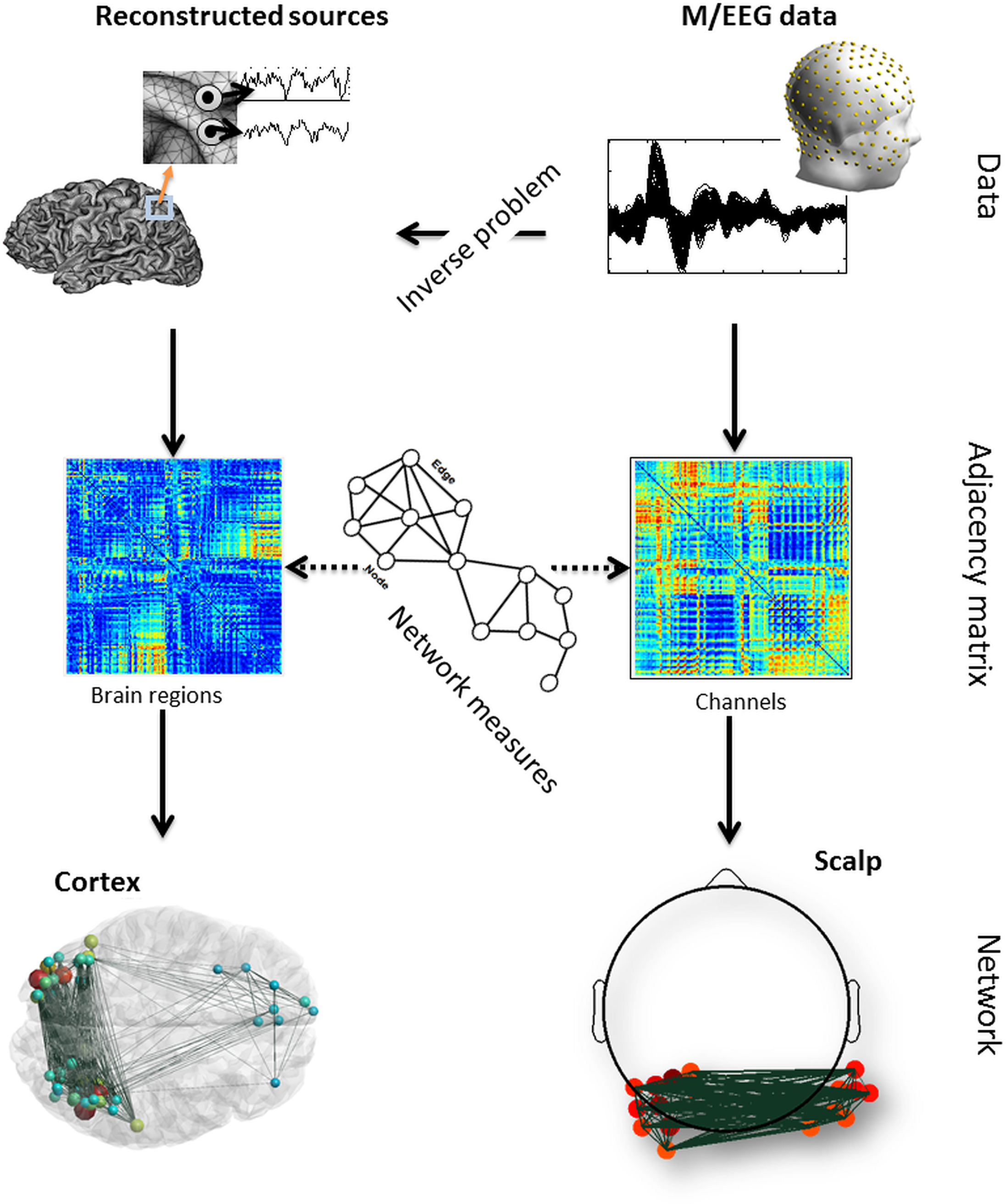
The brain is a large-scale complex network often referred to as the “connectome”. Exploring the dynamic behavior of the connectome is a challenging issue as both excellent time and space resolution is required. In this context, Magneto/Electroencephalography (M/EEG) are effective neuroimaging techniques allowing for analysis of the dynamics of functional brain networks at scalp level and/or at reconstructed sources. However, a tool that can cover all the processing steps of identifying brain networks from M/EEG data is still missing. In this paper, we report a novel software package, called EEGNET, running under MATLAB (Math works, inc), and allowing for analysis and visualization of functional brain networks from M/EEG recordings. EEGNET is developed to analyze networks either at the level of scalp electrodes or at the level of reconstructed cortical sources. It includes i) Basic steps in preprocessing M/EEG signals, ii) the solution of the inverse problem to localize/reconstruct the cortical sources, iii) the computation of functional connectivity among signals collected at surface electrodes or/and time courses of reconstructed sources and iv) the computation of the network measures based on graph theory analysis. EEGNET is a unique tool that combines the M/EEG functional connectivity analysis and the computation of network measures derived from the graph theory. The first version of EEGNET is easy to use, flexible, and user friendly.